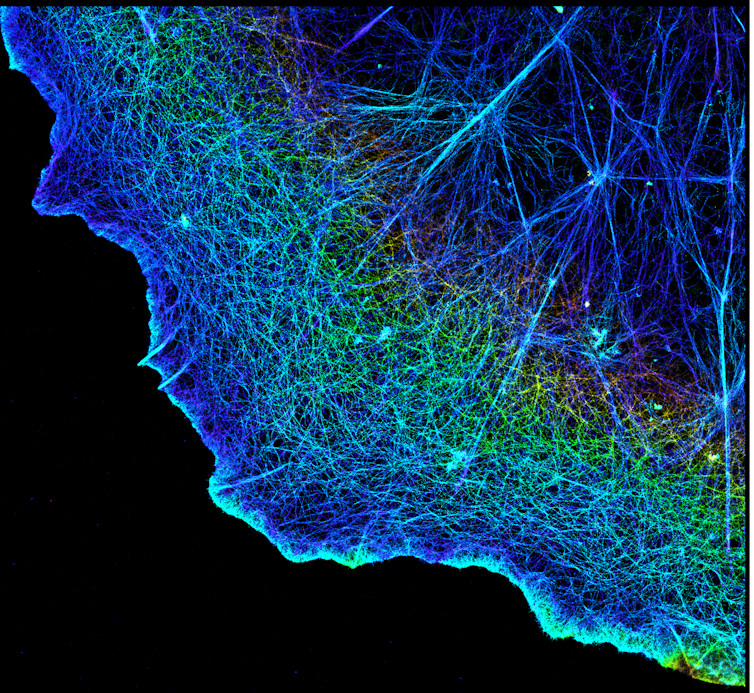
Cell division, or the process of how daughter cells emerge from a mother cell, is fundamental to biology. Every cell inherits the same protein and DNA building blocks that make up the cell it originally came from. Yet exactly how these molecular building blocks arrange themselves into new cells has remained a mystery.
Studying cell division requires simultaneously viewing nanometer-scale macromolecules like proteins and DNA all the way up to millimeter-scale populations of cells, and over a time frame that ranges from seconds to weeks. Previous microscopes have been able to capture tiny objects only in short time frames, typically just tens of seconds. There hasn’t been a method that can examine a wide range of size and time scales all at once.
My team and I at the University of Michigan’s Bioplasmonics Group developed a new kind of superresolution imaging that reveals previously unknown features of how cells divide.

Advancing superresolution imaging
It wasn’t possible to view cells at the molecular level until recently with the 2014 Nobel Prize-winning development of superresolution.
Traditional light microscopes blur very small objects that are close together in a sample, because light spreads out as it moves through space. With superresolution, fluorescent probes attached to the sample could be switched on and off like twinkling stars on a clear night. By collecting and combining many images of these probes, a superresolution image can bring very small objects into view. Superresolution opened a whole new world in biology, revealing structures as small as 10 nanometers, which is about the size of a protein molecule.
However, the fluorescent probes that this technique relies on can quickly wear out. This limits its use in studying processes that take place over extended periods, such as cell division.
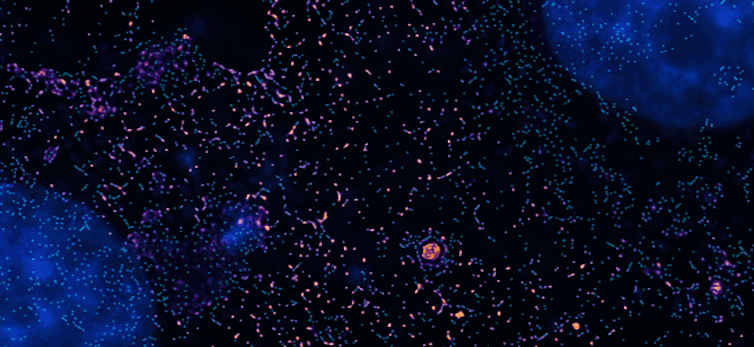
My research team and I have a developed a solution we call PINE nanoscopy. Instead of absorbing light as traditional fluorescent probes do, the probes we use scatter the light so they do not break down with repeated light exposure.
To resolve very small objects that are close together, we built filters made of thin layers of polymers and liquid crystals that allow for detection of scattered light, which triggers the probes to switch on and off. This allowed us to see nanometer-scale details of cells that would otherwise be blurred by traditional microscopes.
Remarkably, we found that these nanometer-scale details could be viewed for very long periods – over 250 hours. These details would typically be lost over time with traditional superresolution methods.
Shedding new light on cell division
We then applied our method to study how molecular building blocks organize in cell division.
We focused on a protein called actin that helps maintain cell structure, among many other functions. Actin is shaped like branching filaments, each about 7 nanometers (millionths of a millimeter) in diameter, that link together to span thousands of nanometers. Using PINE nanoscopy, we attached scattering probes to actin to visually follow human cells as they divided.
We made three observations on how actin building blocks organize during cell division. First, these molecular building blocks expand to increase their connections to their neighbors. Second, they also draw closer to their neighbors to increase their points of contact. And third, the resulting networks tend to contract when the actin molecules are more connected to one another and expand when they are less connected to one another.
Based on these findings, we were able to discover new information about the process of cell division. We found that interactions between actin building blocks sync up with the contraction and expansion of the whole cell during division. In other words, the behavior of the actin molecules is connected to the behavior of the cell: The cell contracts when the actin expands, and it expands when the actin contracts.
Uncovering disease with superresolution
We plan to use our method to study how other molecular building blocks organize into tissues and organs. Like cells, tissues and organs are organized in a hierarchy that can be examined from a scale of small to large. Examining the dynamic and complex process of how protein building blocks interact with one another to form larger structures could advance the future creation of new replacement tissues and organs, such as skin grafts.
We also plan to use our imaging technique to study how protein building blocks become disorganized in disease. Proteins organize into cells, cells organize into tissues and tissues organize into organs. A very small change in building blocks can disturb this organization, with effects that can lead to diseases like cancer. Our technique could potentially help researchers visualize and, in turn, better understand how molecular defects in tissues and organs may develop into disease.
Somin Lee, Assistant Professor of Electrical & Computer Engineering, Biomedical Engineering, University of Michigan
This article is republished from The Conversation under a Creative Commons license.









 Before the Bus Arrives
Before the Bus Arrives 

 Stretch – After hours of sitting, stretching from head to toe can be a good way to get physically active if you’re short on time and stuck at your desk. Neck, shoulder, arm, back and leg stretches are all easy ways to stand up and disengage from the computer. Another trick: practice wrist exercises to avoid injury or strain from typing.
Stretch – After hours of sitting, stretching from head to toe can be a good way to get physically active if you’re short on time and stuck at your desk. Neck, shoulder, arm, back and leg stretches are all easy ways to stand up and disengage from the computer. Another trick: practice wrist exercises to avoid injury or strain from typing.